The mRNA expression experiments were completed at the end of 2010.
Gene expression can be analyzed in our Colonomics Browser
QC analysis
Two samples (one tumor and its paired mucosa) were hybridized in triplicate (one in each plate assay) to assess technical reproducibility of the expression data.
Results show very good quality with small inter-assay variability. See some plots here.
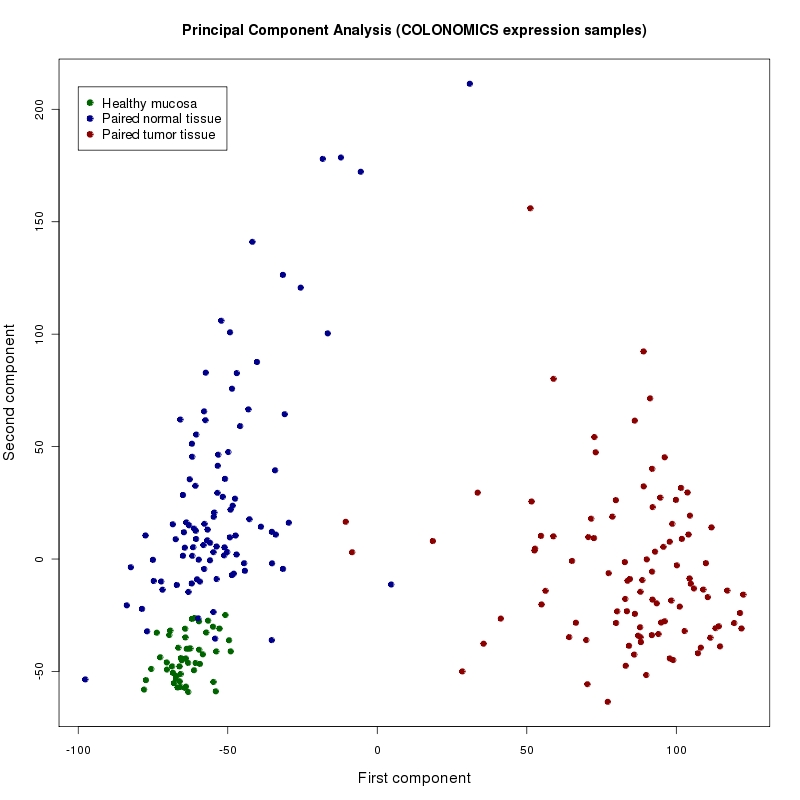
Exploratory analysis
A principal components analysis shows good separation of the samples. This indicates that a large number of differentially expressed genes exist.
Regulatory Networks
Transcriptional regulatory programs have an essential role in cancer.
Our aim is to characterize the differences between transcriptional programs of normal and tumor colon cells, through a reverse engineering reconstruction of gene regulatory networks.
Transcriptional networks for both normal and tumor samples were built using the ARACNe algorithm through gene expression profiles from 200 colon cancer cases (100 tumor and 100 normal adjacent mucosa).
Inference of gene regulatory networks at the whole-genome level has allowed us to detect a generalized loss of transcriptional activity in colon tumors, which had not been described before. Notably, there are also specific transcription factors that hugely increase their number of target interactions in the tumor network.
See additional information here.
Raw data
Expression data are public available at GEO GSE44076